Scientific background and aims of the SFB Fusarium project
F. graminearum is a world-wide ocurring pathogen that is able to colonize numerous host plants without following classical gene-for-gene interactions. This fungus is a threat for food safety since it produces mycotoxins during plant infection, thereby contaminating feed and food. Among the genetic resources of crop plants that are empirically used by plant breeders, only polygenically inherited quantitative differences in resistance exist. It remains largely unknown how Fusarium achieves to escape recognition on so many different host plants. Theoretically, with hundreds of secreted proteins, if a single one is recognized by a matching plant disease resistance gene product, this should lead to avirulence of the fungus and to gene-for-gene type interactions. Since there is no evidence for this on any host, we postulated another twist in the generally accepted ZIG-ZAG scheme, which has been originally proposed by Jones and Dangl.
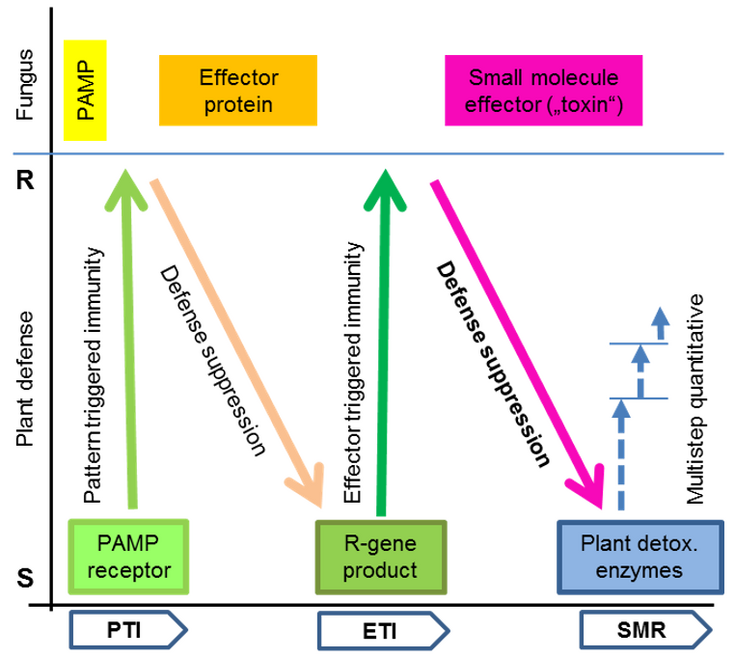
Fig. 1: In the extended ZIG-ZAG model the strength of the plant defense response is indicated by the Y-axis, ranging in the outcome from S (susceptible) to R (resistant). Fungal components affecting the interaction are shown on top assuming the simplest (unrealistic) scenario that a single PAMP (pathogen associated molecular pattern), effector protein or small molecule effector is involved. The plant response can be divided in the classical phases (along X-axis): PTI – pattern-triggered immunity, ETI – effector-triggered immunity and the new addition, SMR – small molecule resistance. SMR is mediated by multiple mechanisms and enzymes (symbolized by dashed arrows) and is effective only to provide a variable intermediate level of resistance. If a second small molecule effector is suppressing defense and is not neutralized, the plant is highly susceptible. For the originally proposed ZIG-ZAG model of the plant immune system see Jones and Dangl (2006): Nature 444, 323.
In innate immunity the first layer consists of recognition of evolutionary conserved molecular pattern of microbes by plant surface receptors. The basal immunity (pathogen associated molecular pattern (PAMP) triggered immunity, PTI) is in general overcome by effector proteins. In case of fungi these effector proteins are delivered inside the plant cell by mechanisms that are hardly understood, and interfere with defense gene expression. Plants can regain resistance by developing intracellular receptors, which trigger a strong defense response when recognizing a corresponding effector protein (ETI – effector triggered immunity).
A major working hypothesis of the SFB is that Fusarium cannot avoid being recognized on so many hosts but is rather able to suppress ETI (and PTI) using multiple, partly redundant small molecule effectors. Our work in the SFB is therefore focused on secondary metabolites, putatively acting as small molecule effectors in Fusarium-plant interaction. The way how plants can cope with this is to develop resistance to such metabolites in various mechanistically different ways such as efflux by transporters of different classes, chemical modification of the fungal metabolite in different ways (e.g. acylation, glycosylation, glutathione conjugation) and sequestration in vacuole, or transport to the apoplast and incorporation into cell wall material. This scenario is consistent with the observed quantitative resistance differences that are polygenically inherited.
The main goal of the SFB is to apply the rapidly evolving tools of genomics, transcriptomics and metabolomics to get a deeper understanding of the molecular mechanisms underlying fungal virulence and plant resistance. This should allow to target fungal virulence mechanisms and to utilize the gained knowledge to solve the Fusarium problem by knowledge based resistance breeding.
