Glyco-Cell Biology
Overview
Overview
Protein localisation signals and mechanisms in plant cells, intracellular transport of glycosylation enzymes and secretory proteins, structure-function of the plant Golgi apparatus, high-resolution optical imaging of live cells
Leader: Ass.Prof. Mag. Dr. Jennifer Schoberer
Research Interests
- Subcellular trafficking of secretory proteins in plant cells
- Subcellular trafficking, dynamics and interactions of plant glycosylation enzymes in the endoplasmic reticulum and Golgi apparatus
- Cell biology of the plant Golgi apparatus
- Development of new approaches to control and modify the glycans on recombinant proteins
- Development of new advanced imaging technologies required for protein traffic research
Orientation
Research in our group concentrates on getting a better understanding of the plant secretory pathway, which serves as a production platform for proteins and polysaccharides. The overall goal is to understand the fundamental cell biology of the Golgi apparatus, which is a central biosynthetic organelle of the secretory pathway in all eukaryotes. With our research we aim to (1) elucidate the organisation of Golgi-resident glycosylation enzymes, (2) determine the signals and underlying mechanisms responsible for organising these enzymes, and (3) use this knowledge to modulate protein glycosylation for various biotechnological applications.
In the centre of our studies is the application of well-established glycosylation enzymes combined with cutting-edge non-invasive bioimaging techniques (e.g. FRET-FLIM, photobleaching) that allow to study the subcellular localisation, transport and interaction of proteins in real-time in living cells at unprecedented resolution and speed. We use the model plants Nicotiana benthamiana and Arabidopsis thaliana and further combine classic cell biological methods with biochemical analysis, genetic tools, and glycosylation analysis.
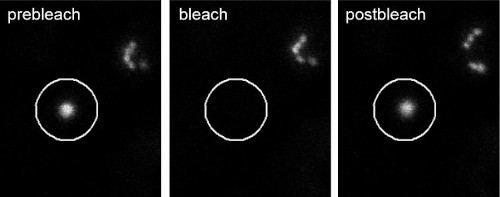
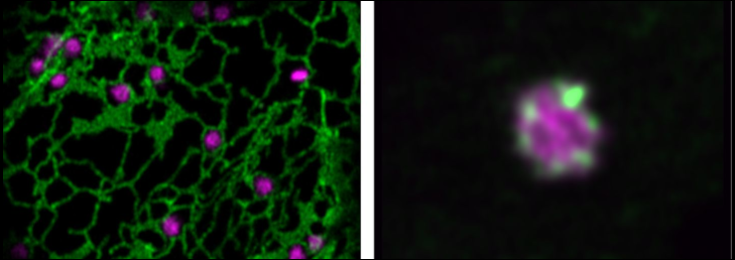
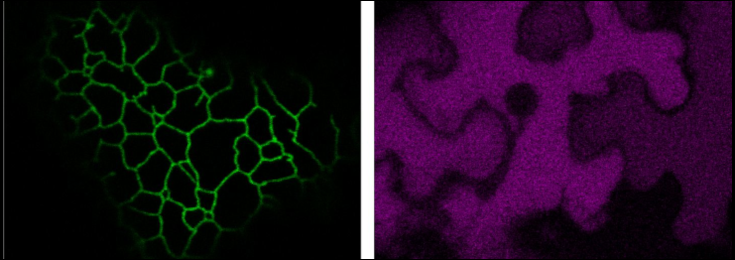
Photobleaching (FRAP) of a single Golgi stack in N. benthamiana leaf cells.
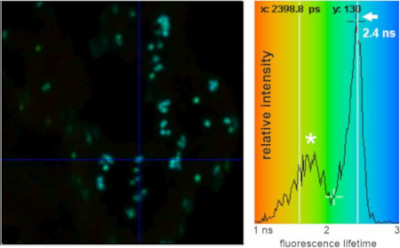
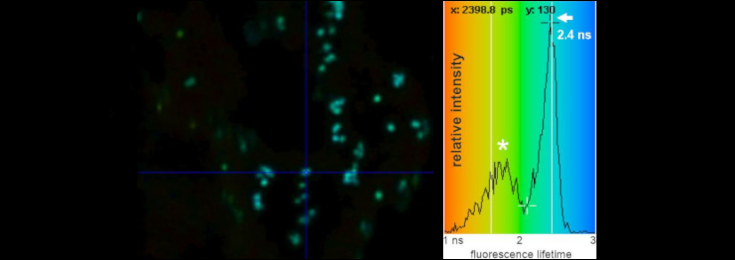
FRET-fluorescence lifetime (FLIM) image of Golgi bodies in N. benthamiana leaf cells highlighted via expression of a GFP-labelled Golgi-resident N-glycan processing enzyme.
Projects
Current Project
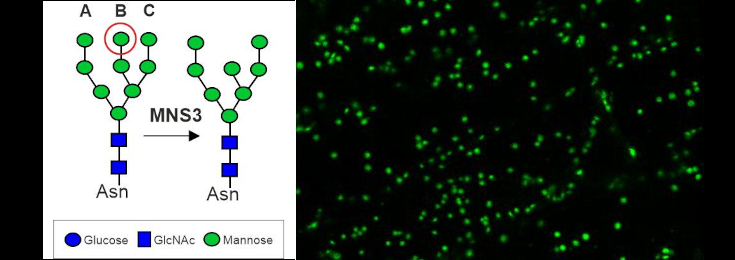
Dissecting the ER-Golgi interface using Arabidopsis MNS3
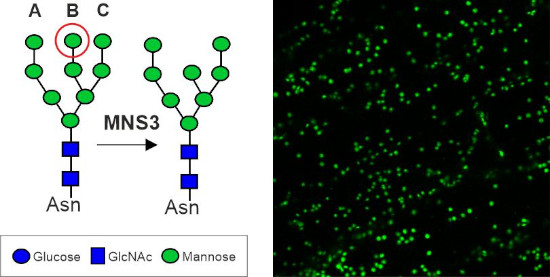
The Arabidopsis MNS3 enzyme fulfils the biosynthetic role of an ER-α-mannosidase that generates oligomannosidic N-glycan structures mainly found on ER-resident glycoproteins, but itself resides in a Golgi compartment that is closely associated with ER exit sites. This project aims to investigate the discrepancy between the proposed in vivo site of action in the ER and putative steady-state localisation of MNS3 in the cis-Golgi.
Publications
Funding Agencies
Collaborations
- Dr. Verena Kriechbaumer, Oxford Brookes University, Oxford, UK
- Prof. Stan Botchway, STFC, Central Laser Facility, Harwell-Oxford, UK