Introduction
Cells and proteins are known to interact with their surroundings not only via chemical or biochemical signals, but also through their ability to sense, transduce and exert (mechanical) forces. Variations in these mechanical properties are indicators of changes in the cellular metabolism or state (e.g. disease, cancer, age,…), and can be used as diagnosis tool in early detection of abnormalities and also be employed to test the efficacy of, for instance, anticancer drug treatments.
Most of our efforts at the Institute for Biophysics are focused on the description of such mechanical response of cells and proteins upon diverse stimuli and the hierarchical contribution of the different cell constituents to the overall performance. Furthermore, it is also known that cellular response, biology and fate highly depend on mechanical features of the underlying substrate. Therefore, a detailed mechanical description of these supports becomes of high relevance. Also proteins, very commonly used as substrate for cell attachment have found their space in this section. For them, the target mechanical factors to determine differ from those calculated for cells, but provide also relevant information on their properties and inner arrangement.
Methods
The evaluation of the mechanical properties of the biological samples are performed with atomic force microscopy (i.e. force spectroscopy, force volume, stress relaxation, and creep measurements). Depending on scale of interest either sharp tips (nano-indentation) or colloidal particles (micro-indentation) are used. Rheological models are employed to interpret the experimental outcome. Homemade software (AFM toolkit) has been programmed in R to simplify data analysis.
Structural features of the biological systems are monitored by atomic force microscopy, confocal fluorescence microscopy, and transmission internal fluorescence microscopy.
Cell and hydrogel mechanics
The study of the mechanical properties of (cancer and healthy) cells connect three research fields, cell biology, biomedicine and physics of materials. In this way, we monitor the response and fate of the cells to external mechanical stress while we advance in the understanding the mechanical and structural function of the different constituents of the cell. Thus, we aim to elucidate mechano-transduction pathways, identification of adhesion mechanisms, and differentiation between healthy and cancerous cells. For these tasks we use different substrate coatings and molecules that originate morphological changes and inner structural rearrangements in response to them (e.g. resveratrol, cytochalasin, etc.).
The understanding of the mechanics of hydrogels complements cell studies. On one hand hydrogels can be used as a rheological simplified model that can be compared to cell complexity. On the other hand, hydrogels can be used as cell scaffolds, and their mechanical properties influences cell attachment, shape, and proliferation.

Left: Cartoon showing a stress relaxation experiment. The force- time curve consists of three segments: approach, pause and retract. The “approach” segment delivers information about molecular and colloidal interactions. The “pause” segment conveys information about the mechanical properties of the cell. The “retract” segment gives information about adhesion and molecular elasticity of cell components.
Right:
(a) Microscopy image of breast cancer cells.
(b) Real force relaxation measurement taken in points (1) and (2) of figure (a).
(c) and (d) High and topography AFM images (a).
Polymer (visco)elasticity
The characterization of polymers from a micrometer to a molecular scale is a line of research that we want to give more importance to in the next three years.
We mainly study proteins. We differentiate between two types of experiments. In the first type, we study the mechanical properties of biomimetic coatings by varying parameters such as temperature, solvent, ionic strength, pH etc. In the second type, we investigate the elasticity of individual proteins as a function of point mutations, solvent polarity, pH, temperature, etc.
Protein coatings are further used for cell attachment studies. We can highlight the use of fibronectin and mucin films. While the former is a protein with selective capability to recognize cell membrane receptors, the latter is a glycoprotein constituting the main component of the mucus secretion of goblet and epithelial cells.
Another interesting example is filamin A (FLNA) which belongs to a family of cytoskeletal actin crosslinking proteins. FLNA is involved in muscle contraction in the cardiovascular system and cardiac health. Furthermore, FLNA mutations in humans seem to cause neuronal and intestinal disorders, while FLNA deficient melanoma cells show severe defects in cell migration. Since FLNA can act as a mechanosensor converting extracellular mechanical signals into intracellular biochemical signals, it is interesting for us to investigate its response to external force.
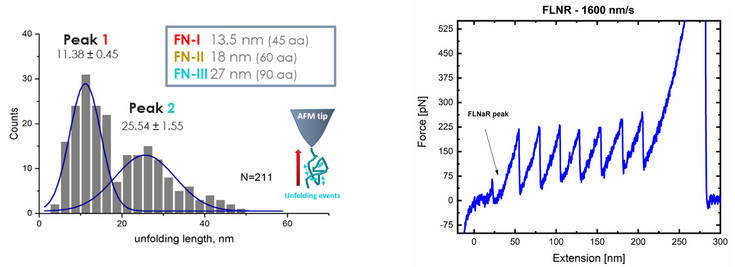
Left: Unfolding lengths of fibronectin domains.
The unfolding force or pattern is obtained from the “retract” segment of the force-distance curve.
The measurement shows that two domains could be unfolded form the fibronectin coating.
Right: Force distance curve of a (I27)3-FLNaR/FLNaQ-(I27)4 hetero-polyprotein. Molecular biology was used to engineer the polyprotein which has seven titin (I27) protein units. The well-known unfolding sawtooth-like pattern of the individual I27 protein domains are used to elucidate the part of the measurement which corresponds to filamin. The small, long peak in the beginning of the measurement is the unfolding of the Filamin A protein.
