Genetic diversity study for Asian and African Zebu cattle and whole genome resequencing for SNP discovery and development of Zebu cattle specific microarray
SUPERVISOR: Johann SÖLKNER
PROJECT ASSIGNED TO: Tafara Kundai MAVUNGA
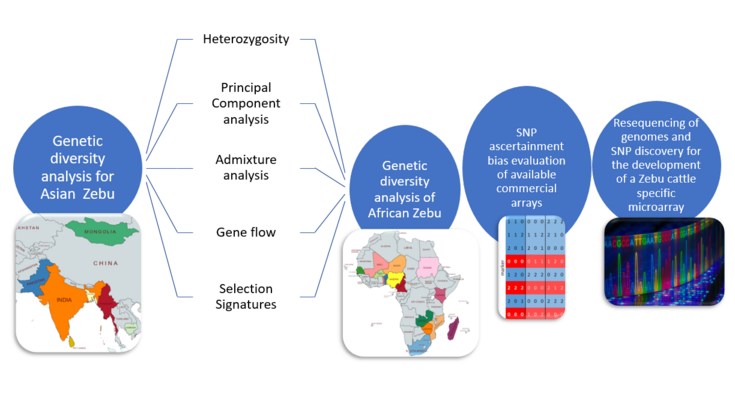
Genetic diversity in cattle is crucial for maintaining the health, adaptability, and overall sustainability of livestock species. Genetic variation within a given population is affected by different processes like mutation, genetic drift, selection, reproductive isolation, and migration, which determine its fitness and adaptation to specific environments. These local cattle breeds harbour favourable characteristics such as adaptation to diverse climatic conditions and disease resistance. The need to characterise these breeds and maintain genetic variability is of utmost importance to ensure that traits which are valuable in certain climates or production systems are preserved. Some breeds are already considered rare or endangered due to declining population numbers hence genetic diversity analyses will help draw direction to conservation efforts. However, the use of available commercial SNP arrays for genetic characterisation of most local breeds is currently not capturing all the genetic variation. In this project we aim to evaluate the ascertainment bias of commercial Taurine SNP arrays in evaluating Asian and African Zebu cattle, resequencing their genomes for SNP discovery and development of a Zebu cattle specific microarray. In addition, we aim to evaluate genomic diversity, genetic admixture, population structure, investigate gene flow (migration events) and detect selection signatures related to their uses, stature, and adaptation in Asian and African Zebu cattle. In a first step, genomic characterisation of 36 Asian Zebu cattle breeds is performed. The general workflow of this PhD is below.