Molecular analysis of beet crop diversification
SUPERVISORS: Juliane DOHM and Heinz HIMMELBAUER
PROJECT ASSIGNED TO: Felix SANDELL
Despite continuous efforts to characterize the genomes of agriculturally important beet crops like table beet, sugar beet, red beet, and chard, the genetic basis for domestication and diversification in the genus Beta is unknown. As of today, only one example of a beet domestication gene has been described. The crops of the genus Beta have strikingly different phenotypes, but are genetically very closely related, complicating the calculation of population structure on the basis of whole genome sequencing data. In previous work, the phylogenetic relationship between different species of wild beets and sugar beet was analysed, however, the phylogenetic relationship among cultivated forms has yet to be explored. It is expected that the genetic distance between cultivated beet crops is much lower than between wild taxa of the genus Beta so that previously employed approaches based on representative MinHash sketches may fail. Therefore, single-nucleotide variants will be assessed to address relevant questions in beet phylogeny, domestication and diversification. Furthermore, clustering and dimensionality reduction techniques will be tested and optimized to identify distinctive features for each cultigroup. In total, I will analyse 1300 genomes of beets that are fully sequenced at low coverage. With this work I hope to further clarify the genetic relationship between beet cultivars and wild beet species and to identify regions (and genes) that play an important role in the process of beet crop domestication and diversification.
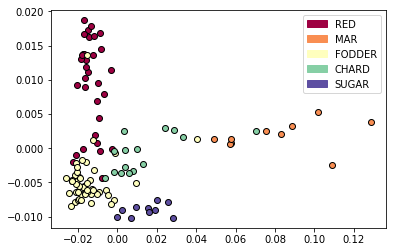
Figure 1: Clustering of Beta accessions based on MASH distances
