HISTOVAR: Role of the histone variant H2A.Z in pathogenic Fusarium species
SUPERVISORS: Lena STUDT and Joseph STRAUSS
PROJECT ASSIGNED TO: Anna ATANASOFF-KARDJALIEFF
The genus Fusarium is widely distributed and found among the economically most important phytopathogenic fungi worldwide. Fusaria are able to colonize a wide range of host plants, including many crop cultivars. Plant infections not only lead to an enormous yield loss but may result in contamination of food and feed with mycotoxins produced by the pathogen. Fusaria are able to produce a plethora of these natural products also referred to as secondary metabolites (SMs), including hazardous mycotoxins, which pose a serious health threat to humans and animals. For the development of novel, sustainable and most importantly efficient strategies against fungal infections of crop plants, it is necessary to unravel the underlying regulatory network responsible for the expression of these pathogenesis- and SM-related genes.
In eukaryotes, all DNA-dependent processes including gene expression depends on the chromatin structure. Chromatin is the fundamental packaging form of DNA, which is wrapped around an octamer of canonical core histones H2A, H2B, H3 and H4. This structure generates a natural obstacle to the transcription machinery and thus must be highly dynamic. Alterations in chromatin structure involve posttranslational modifications (PTMs), chromatin-modifying enzyme complexes or the exchange of core histones with so-called histone variants. These structural changes allow the fungus to express pathogenesis- and SM-related genes when needed, thereby providing an advantage for the pathogen to overcome host defense mechanisms and to successfully establish the infection within its host.
The project HISTOVAR focuses on chromatin dynamics related to the role of histone variant H2A.Z in the two Fusarium species, F. fujikuroi and F. graminearum, preferably infecting rice and wheat, respectively. The role of H2A.Z in filamentous fungi is still enigmatic and often conflictual functions are reported, even within the same organism. The project is designed as a collaborative project between an Austrian and a French research group who both aim to understand mechanisms involved in pathogenesis and SM production. Through a combination of reverse genetic and whole genome approaches, HISTOVAR will provide knowledge of H2A.Z’s role and function in fungal development, pathogenicity and secondary metabolism.
The collected knowledge, will contribute to the development of novel strategies against crop loss and mycotoxin contaminations of these and likely other plant pathogenic fungi.
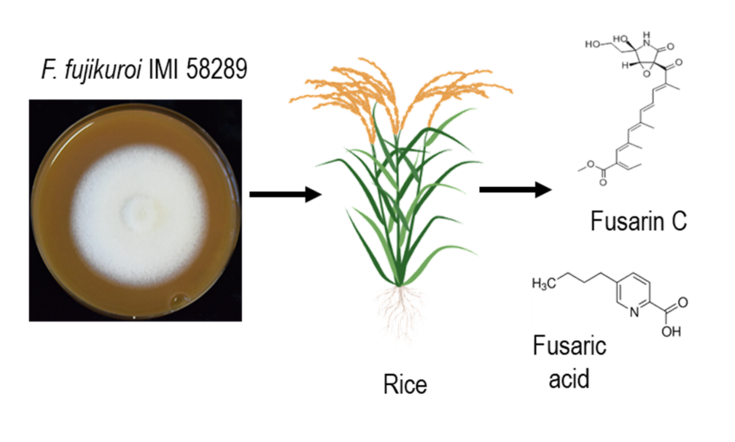
Figure 1: Schemata of rice infection by F. fujikuroi. F. fujikuroi start its fungal attack by the colonization of rice roots. Thereafter, the fungus can gain advantage by producing and releasing mycotoxins. Some classes of these natural substances not only damage the plant they contaminate food and feed, causing serious health issues.
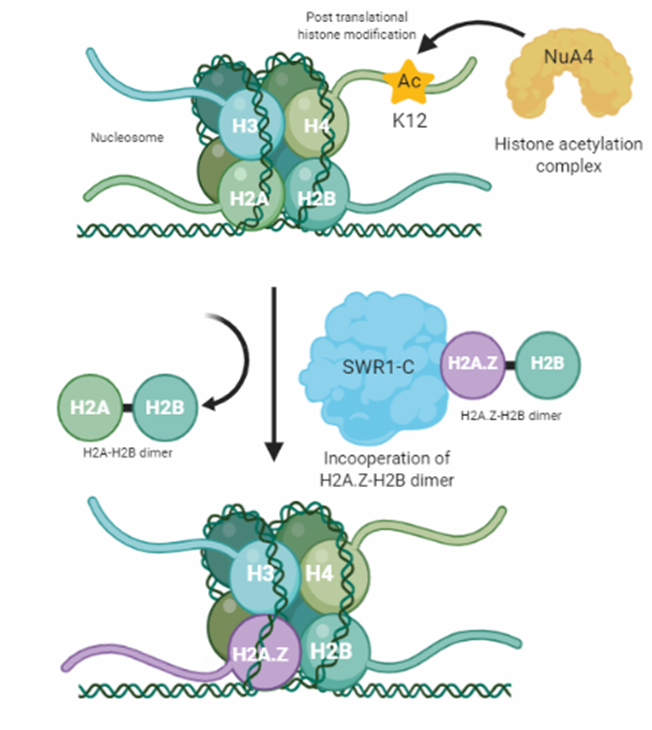
Figure 2: Visualizes the theoretical process of H2A.Z deposition. The process is started by the NuA4-dependent histone 4 lysine 12 acetylation (H4K12Ac). Then, H4K12Ac serves as an anchor and recognition signal for the multi modular SWR1-C protein complex which catalytic subunit Swr1 exchanges a H2A-H2B histone dimer against a H2A.Z-H2B dimer, facilitating H2A.Z deposition.
