In-silico bioprospecting of novel enzymes from underexplored habitats
SUPERVISOR: Doris RIBITSCH
PROJECT ASSIGNED TO: Maren RICKERT
The development of bio-based polymers with unique chemical functionalities will be highly significant in future in order to reduce the dependence on fossil fuels and emission of carbon dioxide. Bio-based compounds can be obtained through selective transformation of building blocks originating from plants and other non-fossil, biogenic feedstocks for replacement of fossil carbon feedstocks. Ligninocellulose-based and -reinforced biopolymers have gained particular attention. However, access to well-defined, bio-based polymers requires the development of efficient depolymerization strategies, for which enzymes have a particularly high potential due to their high specificity and selectivity. At the same time, such enzymes can be used to predict the biodegradability of new polymers in different environments.In this PhD thesis, functional screening of underexplored habitats such as anaerobic fungi and archaea as well as strain collections isolated from selected environments such as wastewater treatment plants will be performed to discover new enzyme activities. The responsible enzymes will be identified and isolated. Rapid high-throughput sequencing of whole genomes and metagenomes will be applied to explore sequences by computational tools for identification of novel enzymes, also known as in-silico bioprospecting, which has come up as an efficient, cost and time effective method that also allows identification of enzymes even from non-cultivable strains. Finally, the identified sequences will be produced in expression hosts like E.coli and P.pastoris, purified and characterized in term of biochemical and kinetic parameters, substrate specificities and reaction mechanisms.
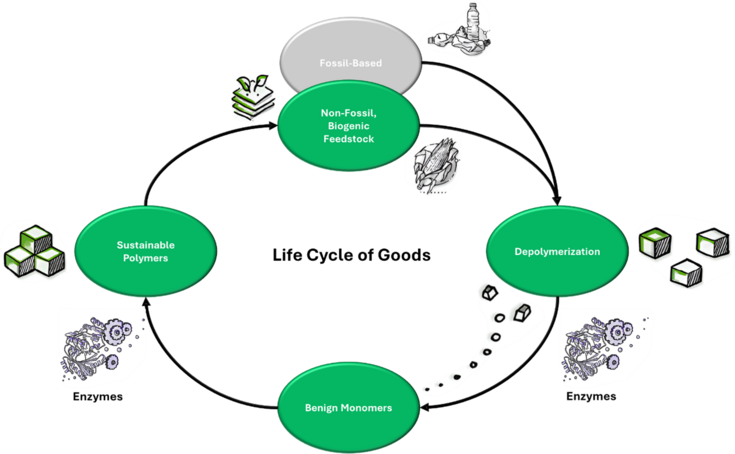
Fig1: Schematic of a circular polymer economy in which the discovery of novel enzymes enables the efficient depolymerization of diverse feedstock into benign monomers, which can be reused for synthesis of sustainable polymers.
