PolyBacTex: Converting mixed textile waste into recycled fiber
SUPERVISOR: Diethard MATTANOVICH
PROJECT ASSIGNED TO: SUKSHAM
In alignment with the circular economy strategy, the development of sustainable textile recycling techniques that effectively treat blended textile waste would lead to a reduction in the incineration of textile waste.
The aim of the project is to recycle mixed textile waste by means of alkaline hydrolysis of polyethylene terephthalate (PET) into the monomers ethylene glycol (EG) and terephthalic acid (TPA) and utilize these monomers as carbon sources to ferment cellulose-producing bacteria.
The initial phase of the project focuses on optimization/intensification of the alkaline hydrolysis process[1] of mixed textile waste to recover cotton fibers and degrade the polyester fraction. Furthermore, the characterisation of metabolic pathways to convert EG and TPA into bacterial cellulose[2] is required to identify the key enzymes to be used for successful strain engineering.
A major part of the project is to modify the cellulose-producing bacteria Komagataeibacter spp. via genetic engineering using synthetic biology tools, such as Golden Gate Cloning. Adaptive laboratory evolution (ALE) will further be used to improve the uptake of monomers and enhance the overall yields of bacterial cellulose production.
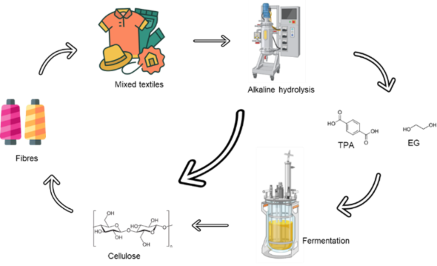
Figure 1- Schematic representation of the project workflow

Figure 2- Linking adaptive laboratory evolution and synthetic biology to generate high performance mutant
[1] Palme, A., Peterson, A., de la Motte, H., Theliander, H., & Brelid, H. (2017). Development of an efficient route for combined recycling of PET and cotton from mixed fabrics. Textiles and Clothing Sustainability, 3(1), 4. doi.org/10.1186/s40689-017-0026-9
[1] Esmail, A., Rebocho, A. T., Marques, A. C., Silvestre, S., Gonçalves, A., Fortunato, E., Torres, C. A. V., Reis, M. A. M., & Freitas, F. (2022). Bioconversion of Terephthalic Acid and Ethylene Glycol into Bacterial Cellulose by Komagataeibacter xylinus DSM 2004 and DSM 46604. Frontiers in Bioengineering and Biotechnology, 10. doi.org/10.3389/fbioe.2022.853322
