Purification of recombinant secretory immunoglobulin A and its biophysical and biochemical characterization
SUPERVISOR: Rainer HAHN
PROJECT ASSIGNED TO: David SCHEICH
The overarching goal of the project “deCIPHER” is the purification and characterization of secretory immunoglobulin A (sIgA) from CHO cell culture supernatant. sIgA has big potential for passive immunization and other therapeutic applications [1-3], and it is the second most abundant type of antibody in the human body, playing a prominent role in host-pathogen defense [4, 5]. sIgA is a large complex molecule consisting of five different polypeptide chains that must be correctly assembled. Monomeric IgA (mIgA) is formed by linking the heavy chain (HC) and light chain (LC) through disulfide bridges. Dimeric IgA (dIgA) is formed linkage of two mIgA to a J chain (Figure 1).
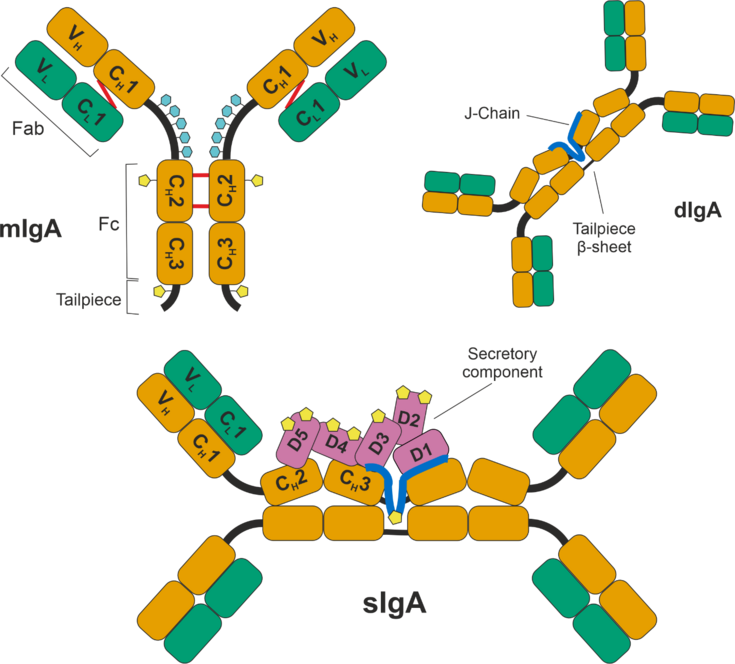
Figure 1: Schematic representation of the structure of monomeric IgA (top left), dimeric IgA (top right) connected via the Joining Chain (J-Chain) and secretory IgA (bottom), including the secretory component (SC). Heavy chains (HC) of the IgA molecules are shown in orange, light chains (LC) in green. mIgA is shown as the IgA1 subtype, which has a longer hinge region with up to 5 O-glycosylation (light blue) sites per chain. N-glycosylation sites are shown in yellow and found on the HC, J-Chain and SC. The J-Chain connects with two of the tailpieces of HC, while the remaining two form a β-sheet. The SC consists of 5 subunits, which reside on one side of the sIgA molecule.
Through association with a secretory component (SC) sIgA is formed [6]. In order to exploit the therapeutic potential of recombinant sIgA expressed in CHO cells, a platform process for capture of the antibody directly from the clarified culture supernatant would be desirable. Immunoaffinity chromatography with a single domain antibody ligand is currently the only option for a capture step directly from the culture supernatant. We envision a downstream platform process featuring a novel biomimetic chromatographic matrix based utilizing Spore coat polysaccharide biosynthesis protein (SpsA) from Streptococcus pneumoniae [7]. SpsA binds specifically to the SC of sIgA [8] and will be immobilized on different bead-based matrices to replace the current the capture step. Different immobilization strategies will be tested and the resulting matrices will be compared and optimized. The capture step will likely be followed by a secondary chromatographic polishing step, focusing on scalability for the process.
Once a reliable downstream processing is established, the project will shift towards characterization of sIgA rather than the purification. The theoretical size of sIgA is 410 kDa, making it one of the larger immunoglobulins. Two subclasses, IgA1 and IgA2, have been identified, which show differences in the constant heavy chains [9]. Moreover, the hinge region of IgA1 is extended and offers additional sites for O- and N-glycosylation [10]. The total amount of possible glycosylation sites is above 30, resulting in a large variation of sIgA variants. Separation and characterization of the different variants under native conditions poses a challenge regarding the conservation of tertiary structure. Analytical methods will involve surface plasmon resonance and glycoanalysis.
Furthermore, secretory IgA (sIgA) DSP and biochemical and biophysical profiling methods to correlate chromatographic retention data with biological function of molecule will be established. Lastly, biomimetic chromatography will be integrated in the MD-LC platform and proof-of-concept will be established for profiling sIgA variants induced by operational effects in a downstream processing workflow, and biochemical properties will be directly linked to biophysical protein characteristics. Upon success, we move closer to unlocking the potential of emerging biopharmaceuticals via comprehensive analysis of drug candidates and to obtain fundamental insights in up- and downstream processing workflows.
Resources
The doctoral research is part of project (I 5640-N, 'deCIPHER'), which is supported by the Austrian Science Fund, FWF. sIgA containing CHO cell culture supernatant and SpsA ligands are provided by Icosagen Cell factory.
- References
[1] P.N. Boyaka, Inducing Mucosal IgA: A Challenge for Vaccine Adjuvants and Delivery Systems, J Immunol 199(1) (2017) 9-16. https://doi.org/10.4049/jimmunol.1601775.
[2] H.A. Parray, S. Shukla, R. Perween, R. Khatri, T. Shrivastava, V. Singh, P. Murugavelu, S. Ahmed, S. Samal, C. Sharma, S. Sinha, K. Luthra, R. Kumar, Inhalation monoclonal antibody therapy: a new way to treat and manage respiratory infections, Appl Microbiol Biotechnol 105(16-17) (2021) 6315-6332. https://doi.org/10.1007/s00253-021-11488-4.
[3] A. Richards, D. Baranova, N.J. Mantis, The prospect of orally administered monoclonal secretory IgA (SIgA) antibodies to prevent enteric bacterial infections, Hum Vaccin Immunother 18(2) (2022) 1964317. https://doi.org/10.1080/21645515.2021.1964317.
[4] C. de Fays, F.M. Carlier, S. Gohy, C. Pilette, Secretory Immunoglobulin A Immunity in Chronic Obstructive Respiratory Diseases, Cells 11(8) (2022) 1324. https://doi.org/10.3390/cells11081324.
[5] A. Hockenberry, E. Slack, B.M. Stadtmueller, License to Clump: Secretory IgA Structure-Function Relationships Across Scales, Annu Rev Microbiol 77(1) (2023) 645-668. https://doi.org/10.1146/annurev-micro-032521-041803.
[6] A. Bonner, A. Almogren, P.B. Furtado, M.A. Kerr, S.J. Perkins, Location of secretory component on the Fc edge of dimeric IgA1 reveals insight into the role of secretory IgA1 in mucosal immunity, Mucosal Immunol 2(1) (2009) 74-84. https://doi.org/10.1038/mi.2008.68.
[7] S. Hammerschmidt, S.R. Talay, P. Brandtzaeg, G.S. Chhatwal, SpsA, a novel pneumococcal surface protein with specific binding to secretory immunoglobulin A and secretory component, Mol Microbiol 25(6) (1997) 1113-24. https://doi.org/10.1046/j.1365-2958.1997.5391899.x.
[8] Y. Wang, G. Wang, Y. Li, Q. Zhu, H. Shen, N. Gao, J. Xiao, Structural insights into secretory immunoglobulin A and its interaction with a pneumococcal adhesin, Cell Res 30(7) (2020) 602-609. https://doi.org/10.1038/s41422-020-0336-3.
[9] J.M. Woof, M.A. Kerr, IgA function – variations on a theme, Immunology 113(2) (2004) 175-177. https://doi.org/https://doi.org/10.1111/j.1365-2567.2004.01958.x.
[10] J.M. Woof, M.A. Kerr, The function of immunoglobulin A in immunity, J Pathol 208(2) (2006) 270-82. https://doi.org/10.1002/path.1877.
