Characterization of HEK293 cell lines for rAAV production
SUPERVISOR: Nicole BORTH, Astrid DÜRAUER
PROJECT ASSIGNED TO: Georg SMESNIK
Modern gene therapy could serve as a corrective intervention to supplement or repair defective genes due to viral vector based delivery of DNA [1]. Among other viral based methods, the most promising approach for gene delivery relies on adeno-associated virus (AAV) vectors [2]. The manufacturing of recombinant therapeutic AAVs utilizes mammalian cell factories, such as HEK cell lines, which may exhibit notable variations in the efficiency of viral particle production [3], while the underlying processes remains poorly understood [4].
As demonstrated in other domains, such as biopharmaceutical antibody production [5], genome-scale studies incorporating reference genome sequences [6], transcriptome and proteome analysis [7], and exploration of epigenetic mechanisms have facilitated progress in identifying and overcoming limiting factors within these fundamental biological processes [8]. Thus, these strategies will be applied to generate a deeper understanding of cellular processes in the context of rAAV production.
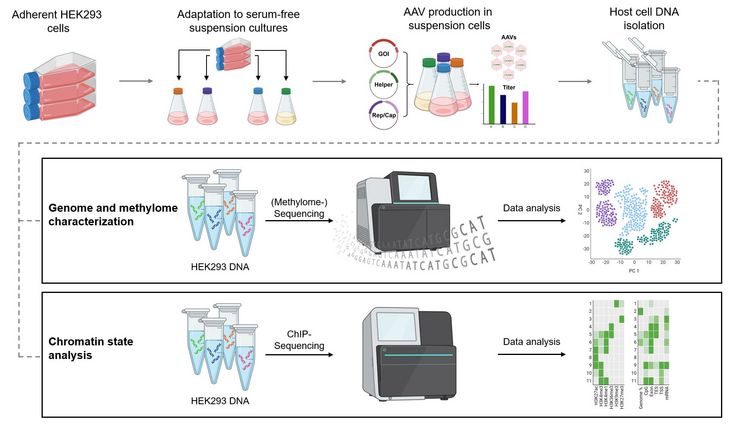
The aim of this PhD thesis is to execute a systematical investigation on rAAV production cell lines derived from adherent HEK293 cells. A comprehensive analysis of the cellular response to various environmental pressures including viral particle production is utilized to identify restricting cellular limitations that contribute to bottlenecks in the production process of rAAVs. The characterization strategy on a genome scale includes investigation of the host cell genome sequences, DNA-methylation patterns, examination of histone modifications and transcriptomic analysis.
For industrial scale-up processes, the preferred conditions require cultivation of production cells in serum-free suspension cultures. To meet this demand, staring from the standard adherently growing HEK293 cell line, a direct adaptation strategy is used to facilitate the shift to serum-free suspension growth in various media compositions. In order to assess the impact of the adaptation process on the genome and epigenome, whole-genome sequencing and methylome sequencing of both the original adherent cell line and the resulting adapted cell lines will be employed. Comparative analysis of these data aims to reveal distinctions between different culture conditions and identify changes arising from the adaptation process.
To address the impact of the cellular response to rAAV production, diverse production scenarios involving various AAV serotypes and different transgene variants will be executed and assessed. Transcriptomic analysis will be utilized to detect changes in transcription patterns, while chromatin immunoprecipitation analysis will be performed to investigate the influence on histone modifications, representing a significant mechanism in epigenetic transcription regulation. Thus, the outcomes of this systematic exploration might pave the way for targeted cell line- and process optimization for efficient production of rAAVs with constantly high yield and quality.
References:
[1] Büning H. Gene therapy enters the pharma market: the short story of a long journey. EMBO Mol Med. 2013 Jan;5(1):1-3. doi: 10.1002/emmm.201202291. PMID: 23283747; PMCID: PMC3569647.
[2] Wang, D., Tai, P.W.L. & Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov. 18, 358-378. doi: 310.1038/s41573-41019-40012-41579. (2019).
[3] Grieger, J.C., Soltys, S.M. & Samulski, R.J. Production of Recombinant Adeno-associated Virus Vectors Using Suspension HEK293 Cells and Continuous Harvest of Vector From the Culture Media for GMP FIX and FLT1 Clinical Vector. Mol Ther. 24, 287-297. doi: 210.1038/mt.2015.1187. Epub 2015 Oct 1036. (2016).
[4] Mingozzi, F. & High, K.A. Immune responses to AAV in clinical trials. Curr Gene Ther. 11, 321-330. doi: 310.2174/156652311796150354. (2011).
[5] Stolfa, G. et al. CHO-Omics Review: The Impact of Current and Emerging Technologies on Chinese Hamster Ovary Based Bioproduction. Biotechnol J. 13, e1700227. doi: 1700210.1701002/biot.201700227. Epub 201702017 Nov 201700215. (2018).
[6] Brinkrolf, K. et al. Chinese hamster genome sequenced from sorted chromosomes. Nat Biotechnol. 31, 694-695. doi: 610.1038/nbt.2645. (2013).
[7] Hausmann, R., Chudobová, I., Spiegel, H. & Schillberg, S. Proteomic analysis of CHO cell lines producing high and low quantities of a recombinant antibody before and after selection with methotrexate. J Biotechnol. 265:65-69., 10.1016/j.jbiotec.2017.1011.1008. Epub 2017 Nov 1012. (2018).
[8] Feichtinger, J. et al. Comprehensive genome and epigenome characterization of CHO cells in response to evolutionary pressures and over time. Biotechnol Bioeng. 113, 2241-2253. doi: 2210.1002/bit.25990. Epub 22016 Apr 25929. (2016).
