Development of high titre soluble expression systems for recombinant protein expression in Escherichia coli via the application of solubility tags and the utilisation of phage derived 5’ UTR expression enhancers
SUPERVISOR: Gerald STRIEDNER
PROJECT ASSIGNED TO: Christoph KÖPPL
Around 30 % of all approved biopharmaceuticals are produced in E. coli (Baeshen et al., 2015). However, complex proteins are often expressed with insufficient titres, in inclusion bodies or in inactive and misfolded forms (Rosano and Ceccarelli, 2014). One way to circumvent these problems is to express challenging proteins in combination with a fusion partner, which often enables the target protein to overcome these challenges (Esposito and Chatterjee, 2006).
In this doctoral thesis, a fusion tag will be developed to enable the soluble production of relevant biopharmaceuticals. Furthermore, a 5’ untranslated region (UTR) expression enhancer will be developed and combined with the solubility tag to further boost the expression level of the target proteins. This expression enhancer – solubility tag combination should be applicable to a wide range of recombinant biopharmaceuticals, providing a step towards a universally applicable UTR - fusion tag combination. In contrast to a great number of already existing fusion tags for recombinant protein production, this system will feature a relatively short fusion tag to keep the exerted additional stress on the cellular transcription machinery on a minimal level. The resulting system will be evaluated in a series of laboratory scale fed-batch fermentations, where certain process parameters will be adjusted to obtain an ideal production process for each model protein.
However, fusion tags are generally unwanted on the therapeutic target protein after purification because they can lead to an unfavourable immune response of the patient (Cserjan-Puschmann et al., 2020). Therefore, this doctoral project will be carried out in cooperation with downstream processing, where the cleavability of the fusion tag with a protease-based cleavage system will be evaluated. This will yield the finished active biopharmaceutical with a native N terminus.
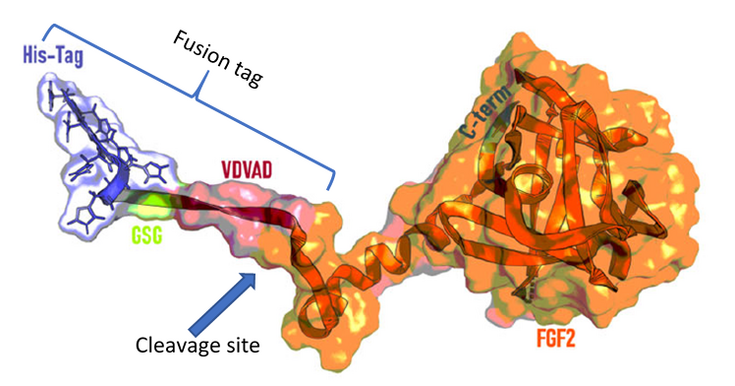
Figure 1. Structure of hFGF2 (Lingg et al., 2021) with N-terminal fusion tag and protease cleavage site on its C-terminus
The results of this doctoral thesis will provide a valuable system to enhance the expression and solubility of recombinant proteins, further establishing the applicability of E. coli as production host for the manufacturing of challenging biopharmaceuticals.
References
BAESHEN, M. N. et al. (2015): Production of Biopharmaceuticals in E. coli: Current Scenario and Future Perspectives. J. Microbiol. Biotechnol. 25: 953-62.
CSERJAN-PUSCHMANN, M. et al. (2020): Production of Circularly Permuted Caspase-2 for Affinity Fusion-Tag Removal: Cloning, Expression in Escherichia coli, Purification, and Characterization. Biomolecules 10.
ESPOSITO, D.; CHATTERJEE, D. K. (2006): Enhancement of soluble protein expression through the use of fusion tags. Current Opinion in Biotechnology 17: 353-358.
LINGG, N. et al. (2021): Advanced purification platform using circularly permuted caspase‐2 for affinity fusion‐tag removal to produce native fibroblast growth factor 2. Journal of Chemical Technology & Biotechnology.
ROSANO, G. L.; CECCARELLI, E. A. (2014): Recombinant protein expression in Escherichia coli: advances and challenges. Frontiers in Microbiology 5.
SWARTZ, J. R. (2001): Advances in Escherichia coli production of therapeutic proteins. Current Opinion in Biotechnology 12: 195-201.
