Host cell response to antibody fragment production in E. coli with special focus on transcriptome and translatome
SUPERVISOR: Gerald STRIEDNER
PROJECT ASSIGNED TO: Sophie Anna VAZULKA Antibody-derived fragments, such as Fabs (Fragment, antigen binding), represent a new and promising class of biopharmaceuticals, due to their capacity to bind specific targets, while allowing for greater tissue penetration than whole antibodies. Compared to antibodies, Fabs show reduced complexity (i.e. no glycosylation), which enables production in E. coli, as an attractive alternative to mammalian cell culture processes. However, there is no generally applicable course of action for microbial Fab production available yet and a “trial and error” approach is needed for each individual Fab. Owing to the adverse effects this class of proteins has on host cell biology and the specific requirements of these proteins to be properly processed and folded, stable and biologically active, their production is a challenging task.
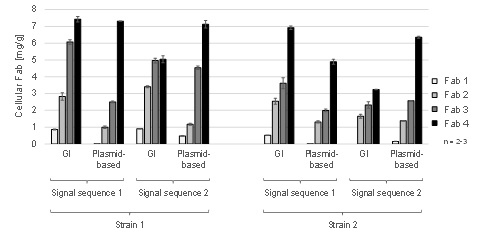
This PhD thesis is conducted in the framework of the CD laboratory for the production of next-level biopharmaceuticals in E. coli, which follows an integrative approach of molecular biology, up- and downstream processing. Model Fabs with high variation in yields and industrially relevant production strains were selected for this study. Plasmid-free T7-based production systems (GI) with site directed integration of a single copy of the gene of interest into the host genome are employed, in order to reduce complexity by reducing metabolic load and plasmid-mediated variation in gene dosage. Different combinations of Fab molecules, periplasmic leader sequences and host strains will be selected and compared.
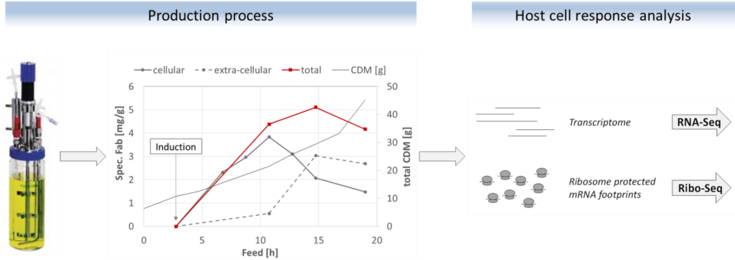
This thesis should help to provide an understanding on host response to periplasmic expression of different Fabs in E. coli on a cellular level, by using Next Generation Sequencing. While mRNA abundances captured by RNA-Seq will be used for transcription profiling, ribosome profiling generates a genome-wide snapshot of translation through sequencing of ribosome protected mRNA fragments. By analysing differential gene expression through the course of defined production processes we aim at characterising the influence of recombinant Fab expression on global transcription patterns and translation in the host cell. The generated knowledge may be used to identify targets for metabolic engineering and optimizing process parameters.