Process development of an integrated continuous two-stage chemostat manufacturing platform using growth-decoupled Escherichia coli
SUPERVISOR: Gerald STRIEDNER
PROJECT ASSIGNED TO: Florian SIMON
Microbial processes are important for the production of a wide variety of products like Fab fragments for medicine (1), enzymes for chemical processes, amino acids (2) and even for the production of alternative protein sources (e.g., precision fermentation to produce cheese (3)). However, there are some major limitations for an ecofriendly long-time production of those molecules. With the current state of the art, continuous microbial protein production processes are not possible due to the high mutation rate of bacteria. Another problem is the high energy and water consumption of large-scale biotechnological plants.
In general, continuous bioprocesses significantly reduce the size of a facility while increasing efficiency (4). However, there are currently two main bottlenecks for the implementation of continuous bioprocesses of bacteria. First, due to the genetic instability (5) of the cells, cultivations under production conditions for more than 3 generations are not feasible, preventing the implementation of such concepts. Second, automation and process control are already a constant challenge in batch-based processes, but they play an even greater role in continuous processes. For a fully integrated continuous process every unit operation must be described by a process model which can anticipate how changes in one of the inputs would migrate through all the unit operations. Although such digital twins are already routinely applied for other industries, there are currently only a few attempts to apply digital bioprocess twins for biotechnological production. Hence, as of today, worldwide, not a single fully integrated continuous bioprocess in manufacturing scale is established applying microbial cells for the production of recombinant proteins.
The main objective of this thesis is to develop a microbial, continuous production process and embed the process into a digital environment. In the process "digital twins" are generated by applying data-driven as well as mechanistic models, so-called hybrid models, as well as fluid-dynamic models. The idea is to use the patented enGenes-eX-Press technology, which uses a growth decoupled expression system, to overcome the problem of process instability and apply it in a two-stage growth-decoupled chemostat bioprocess (6). The first reactor (R1, seed reactor) will be used to produce biomass which is then transferred to the second reactor (R2, production vessel), where production of the POI is induced. By applying the concept of intensified design of experiments (iDoE), intra-experimental shifts of predefined Critical Process Parameters (CPPs) will be used to characterize a whole design space ideally within one bioprocess, making the accelerated bioprocess characterization more economic and ecologic. The main aim of this study is the investigation of the effect of different dilution rate combinations on the overall productivity of the upstream process. Temperature, pH, DO, and induction strategies will be included in these studies as other key process parameters. The data thus generated will lead to a deeper understanding of the process by revealing the mechanistic relationships between dilution rates and protein yields. Two model proteins will be used in this study, Staphylococcus aureus derived Protein A (SpA) as a representative of secreted proteins, and sfGFP as an intracellularly expressed protein. Based on the CFD models combined with hybrid models, a scale-up of the continuous process should be implemented.
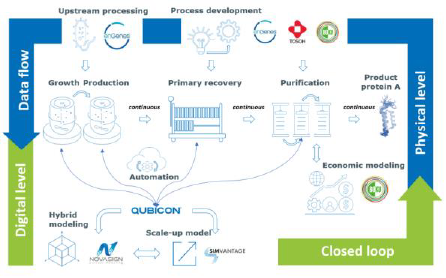
Figure 1: Project overview on ECOnti. The project aims at connecting the physical and the digital world. The connection is implemented a closed loop that allows full control of the developed process design using models (Novasign, SimVantage) and integrative automation (Qubicon). The 2-stage microbial production process is based on an innovation of enGenes and a fully continuous process is developed by enGenes, BOKU and Tosoh
References
1. Better, M., Chang, C. Y., Robinson, R. R., & Horwitz, A. (1988). Escherichia coli Secretion of an Active Chimeric Antibody Fragment. Science, 240(4855), 1041–1043.
doi.org/10.1126/science.3285471
2. Anderhuber, N., Fladischer, P., Gruber-Khadjawi, M., Mairhofer, J., Striedner, G., & Wiltschi, B.
(2016). High-level biosynthesis of norleucine in E. coli for the economic labeling of proteins. Journal of Biotechnology, 235, 100–111. doi.org/10.1016/j.jbiotec.2016.04.03
3. Kang, Y. S., & Richardson, T. H. (1988). Molecular Cloning and Expression of Bovine κ-Casein in Escherichia coli. Journal of Dairy Science, 71(1), 29–40. doi.org/10.3168/jds.s0022-
0302(88)79521-1
4. Yang, O., Qadan, M., & Ierapetritou, M. G. (2019). Economic Analysis of Batch and Continuous Biopharmaceutical Antibody Production: a Review. Journal of Pharmaceutical Innovation, 15(1), 182–200. https://doi.org/10.1007/s12247-018-09370-4
5. Rugbjerg, P., Myling-Petersen, N., Porse, A., Sarup-Lytzen, K., & Sommer, M. O. A. (2018). Diverse genetic error modes constrain large-scale bio-based production. Nature Communications, 9(1). doi.org/10.1038/s41467-018-03232-w
6. Stargardt, P., Feuchtenhofer, L., Cserjan-Puschmann, M., Striedner, G., & Mairhofer, J. (2020). Bacteriophage Inspired Growth-Decoupled Recombinant Protein Production in Escherichia coli. ACS Synthetic Biology, 9(6), 1336–1348. https://doi.org/10.1021/acssynbio.0c00028
